Phages: From Tufts Soil to DNA Sequencing, An ExCollege Research Collaboration

Photo credit: Brendan Carson
Microbiologist Hannah Gavin, PhD, approached the ExCollege in 2019 about offering a special two course sequence on bacteriophages (viruses that infect bacteria), and we jumped at the chance. She developed her phages project at Tufts in conjunction with SEA-PHAGES, an international initiative of the Howard Hughes Medical Institute. In the Fall 2021 ExCollege course, Phages: The Next Frontier in Medicine and Agriculture, the class collected, purified, and characterized novel bacteriophages from soil samples. In the Spring 2022 course, Phages: Microbial Genomics & Bioinformatics, the class is analyzing the DNA sequence data from a subset of those same bacteriophages.
There has been such an enthusiastic response that the Tufts Biology department has agreed to support these courses in subsequent years so that more students can take part in this important research.
We asked Hannah to tell us more about the work her class has been doing and her experience teaching this unique course:
How have students worked with the data gathered during the fall course?
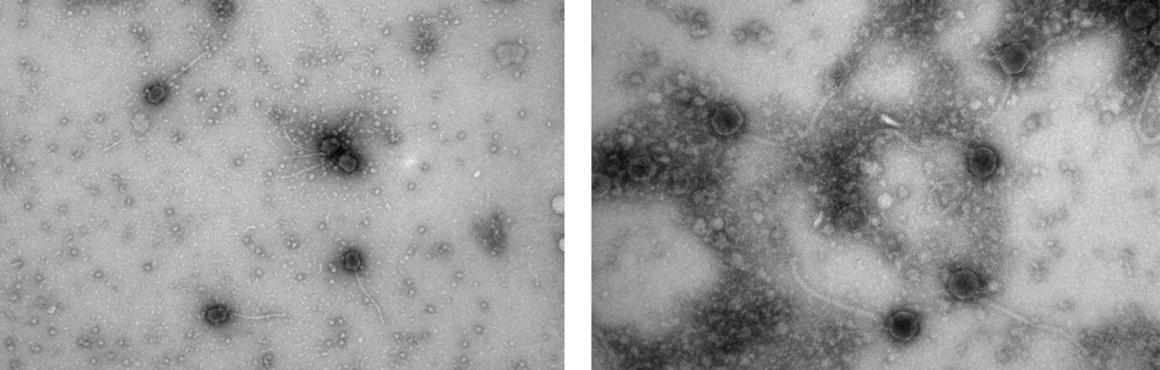
Last semester, eleven students collected soil samples from which we discovered seven new bacteriophages which have now been deposited in a national archive for potential future use (You can view some of these microscopic entities thanks to Transmission Electron Microscopy images courtesy the Harvard Medical School Electron Microscopy facility!) We extracted the genetic material or DNA from two of these phages and, with help from the Pittsburgh Bacteriophage Institute, had it sequenced. That newly sequenced phage genome was the starting point for our current class. First we discussed the process of taking a genome from a physical thing – DNA in a test tube – to data that we can work with and analyze on the computer.
Then, the class split into two teams to complete a process called genome annotation. Annotation involves taking a very close look at the DNA sequence and using bioinformatic tools and databases to determine 1) where genes are located in the genome, and 2) the functions those genes perform. We’ve discussed real-world applications of phages (treatment of antimicrobial-resistant infections, for example) as well as the ways that the specifics of what we are doing with phages connects to bigger concepts in biology.
What has it been like to teach such a seemingly ‘hard science,’ empirical course without asking students to have a background in the biological sciences?
It’s been great. Students are often far more capable of participating in the scientific research process than they realize. From talking with many people over the years, I know there is a perception of a high barrier of entry to science. Partially that comes from the very real complexity of biological phenomena, and partially it arises from other tendencies — the way that field-specific jargon is employed among scientists to expedite communication, for example. To me, the beauty of teaching an empirical “hard science” course with no prerequisites is that we are implicitly acknowledging the intellectual value of every participant, and also explicitly stating that, regardless of their coursework or training up to this point, everyone is going to contribute something valuable to the communal research effort.
Has teaching this class taught you anything about phages? About teaching? About Tufts students?
Ha, it’s taught me so much! Tufts students are awesome. No matter how much I’ve contemplated something we're discussing, someone manages to present or ask something in a way that gives me pause. It’s an endlessly fun challenge to articulate topics clearly, and to deepen my knowledge of topics I’m interested in as both a researcher and as an instructor. It’s a little bit funny that if I’m doing my job well, what we are doing in class should feel achievable, and that’s because it is. But at the same time, it’s also really challenging and impressive. The work that this semester’s students have undertaken represents hundreds of cumulative work hours. The 16 students in my class have analyzed more than 100,000 letters of DNA code and annotated over 160 genes. They’ll collaboratively present two posters at the Tufts 2022 Undergraduate Research and Scholarship Symposium and submit two manuscripts for publication in a journal run by the American Society of Microbiology. My biggest hope is sometime over the coming months or years, each student gets the chance to take a step back from the nitty-gritty of classes, work, and life to say, “wow, we did that!”
The Experimental College wishes to thank Dean Sam Thomas for his support of this course, and the Tufts Biology department, including Michael Grossi and Brendan Carson, for their assistance.